

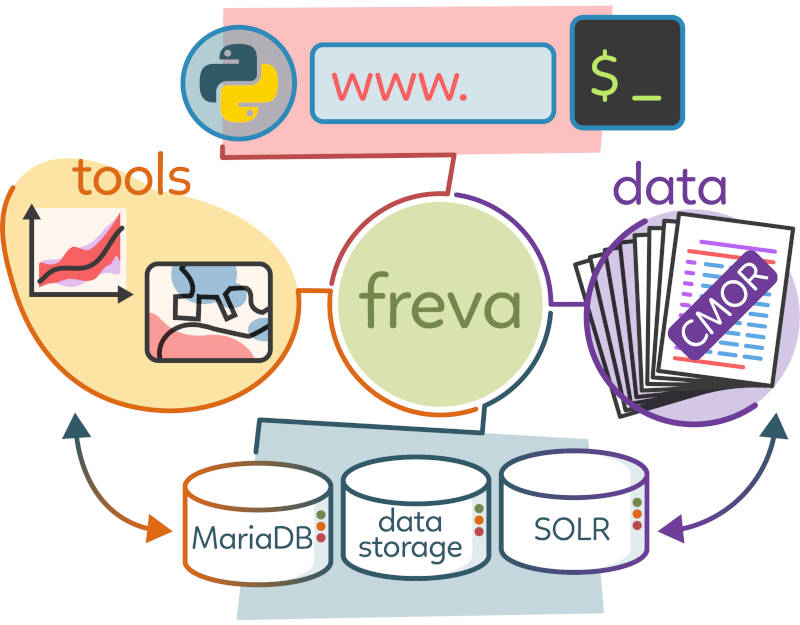
Freva - Data search and analysis framework for the Community
Primer:¶
We will be working with Jupyter Notebooks at the DKRZ Jupyter Hub: Let's head over to https://jupyterhub.dkrz.de/hub/home and start a JHub session
Otherwise, Jupyter notebooks for this workshop: Tutorial-{I,II,III}-*.ipynb
Common Problem: Finding and accesing Data

"I just need 2m-temperature data for my region..."


Sounds familiar?
You're not alone! 🤝
Why should finding data be this hard?
Yet another solution: The Freva framework

Researchers
Need to search
and access data


One stop shop
Adapts to you
Easy to use
Clear process

Perfect for Every Research Task:
Why Choose Freva?
Modellers
Grade
Other Tools
Metadata Store
Smart Architecture:


Setup
The Client Library¶
| Environment | Installation Command |
|---|---|
| modules (Recommended) | module load clint gems |
| conda | conda create -n freva-client-env -c conda-forge freva-client -y |
| pip | pip install freva-client |
Flexible Search
Metadata overview
freva-client databrowser data-overview --host https://www.gems.dkrz.de
Available search flavours: - freva - cmip6 - cmip5 - cordex - nextgems - user Search attributes by flavour: cmip5: - experiment 124 more lines truncated
from freva_client import databrowser
db = databrowser(host="https://www.gems.dkrz.de",
flavour="cmip6",
mip_era="mpi-ge", variable_id="tas")
db
| databrowser(flavour=cmip6, host=https://www.gems.dkrz.de/api/freva-nextgen/databrowser, multi_version=False, mip_era=mpi-ge, variable_id=tas) | |
|---|---|
| # objects | 921 |
| Available search facets for cmip6 flavour | experiment_id, member_id, fs_type, grid_label, institution_id, source_id, mip_era, activity_id, realm, variable_id, time, bbox, time_aggregation, frequency, table_id, dataset, format, grid_id, level_type |
| Available flavours | freva, cmip6, cmip5, cordex, nextgems, user |
Metadata search¶
databrowser.metadata_search(host="https://www.gems.dkrz.de", flavour="cmip6")
{'activity_id': ['aerchemmip',
'afr-22',
'afr-44',
'amsre_soilmoisture',
'amsre_sst',
'arc-44',
'arc-44i',
'arctic_lead_fraction_amsre',
'arctic_lead_fraction_cryosat2',
'arctic_meltponds',
... (9738 more lines truncated) ...
databrowser.metadata_search(host="https://www.gems.dkrz.de", flavour="cmip6").keys()
dict_keys(['member_id', 'experiment_id', 'institution_id', 'source_id', 'activity_id', 'mip_era', 'realm', 'time_aggregation', 'frequency', 'variable_id'])
freva-client databrowser metadata-search --host https://www.gems.dkrz.de
member_id: r001i1850p3, r001i2005p3, r002i1850p3, ... experiment_id: 1pctco2, historical, picontrol, ... institution_id: mpi-m source_id: mpi-esm activity_id: output1 mip_era: mpi-ge realm: atmos, land, ocean, ... time_aggregation: mean frequency: mon, yr, ... variable_id: baresoilfrac, c3pftfrac, c4pftfrac, ... 1 more lines truncated
databrowser metadata-search --host https://www.gems.dkrz.de --facet mpi-ge --flavour cmip6 \
--json | jq -cr 'keys'
activity_id, experiment_id, frequency, institution_id, member_id, mip_era, realm, source_id, time_aggregation, variable_id
What if I don't know the search keys?¶
freva-client databrowser metadata-search --facet mpi-ge \
--host https://www.gems.dkrz.de \
--flavour cmip6
member_id: r001i1850p3, r001i2005p3, r002i1850p3, ... experiment_id: 1pctco2, historical, picontrol, ... institution_id: mpi-m source_id: mpi-esm activity_id: output1 mip_era: mpi-ge realm: atmos, land, ocean, ... time_aggregation: mean frequency: mon, yr, ... variable_id: baresoilfrac, c3pftfrac, c4pftfrac, ...
databrowser.metadata_search("mpi-ge", host="https://www.gems.dkrz.de", flavour="cmip6")
{'activity_id': ['output1'],
'experiment_id': ['1pctco2', 'historical', 'picontrol', '...'],
'frequency': ['mon', 'yr'],
'institution_id': ['mpi-m'],
'member_id': ['r001i1850p3', 'r001i2005p3', 'r002i1850p3', '...'],
'mip_era': ['mpi-ge'],
'realm': ['atmos', 'land', 'ocean', '...'],
'source_id': ['mpi-esm'],
'time_aggregation': ['mean'],
'variable_id': ['baresoilfrac', 'c3pftfrac', 'c4pftfrac', '...']}
Access Metadata¶
freva-client databrowser metadata-search --host www.gems.dkrz.de --flavour cmip6 \
mip_era=mpi-ge variable_id=tas
member_id: r001i1850p3, r001i2005p3, r002i1850p3, ... experiment_id: 1pctco2, historical, picontrol, ... institution_id: mpi-m source_id: mpi-esm activity_id: output1 mip_era: mpi-ge realm: atmos, land, ... time_aggregation: mean frequency: mon variable_id: tas
databrowser.metadata_search(
flavour="cmip6",
mip_era="mpi-ge",
variable_id="tas",
host="www.gems.dkrz.de"
)
{'activity_id': ['output1'],
'experiment_id': ['1pctco2', 'historical', 'picontrol', '...'],
'frequency': ['mon'],
'institution_id': ['mpi-m'],
'member_id': ['r001i1850p3', 'r001i2005p3', 'r002i1850p3', '...'],
'mip_era': ['mpi-ge'],
'realm': ['atmos', 'land'],
'source_id': ['mpi-esm'],
'time_aggregation': ['mean'],
'variable_id': ['tas']}
Counting the number of datasets¶
freva-client databrowser data-count --host www.gems.dkrz.de --flavour cmip6 \
mip_era=mpi-ge frequency=mon variable_id=pr variable_id=tas \
time='2025-01 to 2100-12'
602
db_yr = databrowser(
host="https://www.gems.dkrz.de",
flavour="cmip6",
mip_era="mpi-ge",
frequency="mon",
variable_id="tas",
time="2025-01 to 2100-12"
))
len(db_yr)
602
Accessing dataset paths:¶
freva-client databrowser data-search mip_era=mpi-ge variable_id=tas \
frequency=mon --time "2025-01 to 2100-12" experiment_id=picontrol \
--host https://www.gems.dkrz.de --flavour cmip6
/work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_210001-219912.nc /work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_200001-209912.nc
pcontrol = databrowser(mip_era="mpi-ge",
variable_id="tas",
frequency="mon", time="2025-01 to 2100-12", experiment_id="picontrol",
host="https://www.gems.dkrz.de", flavour="cmip6")
list(pcontrol)
['/work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_210001-219912.nc', '/work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_200001-209912.nc']
Pro Tip: Query data of files¶
freva-client databrowser metadata-search --host www.gems.dkrz.de \
file=/work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_210001-219912.nc
ensemble: r001i1850p3 experiment: picontrol institute: mpi-m model: mpi-esm product: output1 project: mpi-ge realm: atmos time_aggregation: mean time_frequency: mon variable: tas
db.metadata_search(
file="/work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_210001-219912.nc",
host="www.gems.dkrz.de"
)
{'ensemble': ['r001i1850p3'],
'experiment': ['picontrol'],
'institute': ['mpi-m'],
'model': ['mpi-esm'],
'product': ['output1'],
'project': ['mpi-ge'],
'realm': ['atmos'],
'time_aggregation': ['mean'],
'time_frequency': ['mon'],
'variable': ['tas']}
Remote Access
from freva_client import authenticate
import xarray as xr
token = authenticate(host="www.gems.dkrz.de", token_file=Path("~/.token.json").expanduser())
data = databrowser(host="www.gems.dkrz.de", flavour="cmip6",
mip_era="mpi-ge",
variable_id="tas",
experiment_id="historical",
stream_zarr=True)
uri = list(data)[0]
uri
'https://www.nextgems.dkrz.de/api/freva-nextgen/data-portal/zarr/5b8ec334-d2c9-536d-9b8e-249bbd34ac23.zarr'
ds = xr.open_dataset(
uri,
engine="zarr",
decode_times=xr.coders.CFDatetimeCoder(use_cftime=True),
storage_options={
"headers":{"Authorization": f"Bearer {token['access_token']}"}
})
print(ds)
<xarray.Dataset> Size: 138MB
Dimensions: (time: 1872, lat: 96, lon: 192)
Coordinates:
* time (time) object 15kB 1850-01-31 23:52:00 ... 2005-12-31 23:52:00
* lon (lon) float64 2kB 0.0 1.875 3.75 5.625 ... 352.5 354.4 356.2 358.1
* lat (lat) float64 768B -88.57 -86.72 -84.86 -83.0 ... 84.86 86.72 88.57
Data variables:
tas (time, lat, lon) float32 138MB ...
Attributes:
CDI: Climate Data Interface version 1.9.5 (http://mpimet.mpg.de/...
Conventions: CF-1.6
history: Thu Dec 13 08:45:54 2018: cdo -f nc -r setpartabn,tas.parta...
institution: Max-Planck-Institute for Meteorology
CDO: Climate Data Operators version 1.9.5 (http://mpimet.mpg.de/...
.
Freva Databrowser: Hands-on content table
Search
(Findablity)
Find the datasets you need with smart search filters
Cataloging Data
(Reusablity)
Browse, get and share datasets in structured catalogues
Customize Dataset
(Interoperablity)
Add/Remove your customized data to/from Freva
S3 Access
(Accessiblity)
Add and access (your) data via the S3 cloud store
Setup
For this workshop we have two categories of the notebooks.
First and foremost we need to decide to choose one of them. After choosing, please open the Terminal on a new tab on your Jupyterhub session and :
$ module load clint gems
$ da-workshop-setup
Now reload the Jupyterhub page please to make the avaiable kernels "selectable".
And afterward open the Tutorial-py-search-cataloging.ipynb or Tutorial-shell-search-cataloging.ipynb, based on your desire, and then from kernel environment list, please choose, DA Workshop (python) or DA Workshop (shell) based on the chosen language of the notebook.
Now all is set to start! :)
Setup
Initial Check¶
export PATH=/sw/spack-levante/cdo-2.2.2-4z4icb/bin:$PATH
freva-client --version
freva-client: 2508.0.0
from freva_client import databrowser, __version__
print(__version__)
2508.0.0
Search (Findablity)
MPI-GE (Grand Ensemble)¶
Search
$ freva-client databrowser data-overview
Available search flavours: - freva - cmip6 - cmip5 - cordex - nextgems - user Search attributes by flavour: cmip5: - experiment - member_id - fs_type - grid_label - institution_id - model_id - project 118 more lines truncated
Search
databrowser.metadata_search(host="https://www.gems.dkrz.de").keys()
dict_keys(['ensemble', 'experiment', 'institute', 'model', 'product', 'project', 'realm', 'time_aggregation', 'time_frequency', 'variable'])
Search
$ freva-client databrowser metadata-search --host https://www.gems.dkrz.de \
--facet mpi-ge --json | jq -r 'to_entries[] | select(.value[] == "mpi-ge") \
| .key'
project
$ freva-client databrowser metadata-search project=mpi-ge --json \
| jq -rc '.variable | index("tas") != null'
true
Search
metadata = databrowser.metadata_search("mpi-ge", host="https://www.gems.dkrz.de")
facets_with_mpi_ge = [facet for facet, values in metadata.items() if "mpi-ge" in values]
print(facets_with_mpi_ge)
['project']
"tas" in databrowser.metadata_search("mpi-ge",
host="https://www.gems.dkrz.de")["variable"]
True
Search
$ freva-client databrowser metadata-search --host www.gems.dkrz.de --flavour cmip6 \
mip_era=mpi-ge variable_id=tas --time '2025-01 to 2100-12'
member_id: r001i1850p3, r001i2005p3, r002i2005p3, ... experiment_id: picontrol, rcp26, rcp45, ... institution_id: mpi-m source_id: mpi-esm activity_id: output1 mip_era: mpi-ge realm: atmos, land, ... time_aggregation: mean frequency: mon variable_id: tas 1 more lines truncated
Search
from freva_client import databrowser
db = databrowser(host="https://www.gems.dkrz.de",
flavour="cmip6",
mip_era="mpi-ge",
variable_id="tas",
time="2025-01 to 2100-12")
db
| databrowser(flavour=cmip6, host=https://www.gems.dkrz.de/api/freva-nextgen/databrowser, multi_version=False, mip_era=mpi-ge, variable_id=tas, time=2025-01 to 2100-12, time_select=flexible) | |
|---|---|
| # objects | 602 |
| Available search facets for cmip6 flavour | experiment_id, member_id, fs_type, grid_label, institution_id, source_id, mip_era, activity_id, realm, variable_id, time, bbox, time_aggregation, frequency, table_id, dataset, format, grid_id, level_type |
| Available flavours | freva, cmip6, cmip5, cordex, nextgems, user |
Search
Search
$ freva-client databrowser metadata-search --host www.gems.dkrz.de \
--flavour cmip6 mip_era=mpi-ge variable_id=tas --json \
| /run/current-system/sw/bin/jq -c .frequency
["mon"]
$ freva-client databrowser data-count --host www.gems.dkrz.de --flavour cmip6 \
mip_era=mpi-ge frequency=mon variable_id=pr variable_id=tas \
time='2025-01 to 2100-12'
602
Search
db.metadata["frequency"]
['mon']
db_yr = databrowser(
host="https://www.gems.dkrz.de",
flavour="cmip6",
mip_era="mpi-ge",
frequency="mon",
variable_id="tas",
time="2025-01 to 2100-12"
)
len(db_yr)
602
Search
$ freva-client databrowser metadata-search mip_era=mpi-ge variable_id=tas \
frequency=mon --time "2025-01 to 2100-12" --json \
--host https://www.gems.dkrz.de --flavour cmip6 \
| jq -c ".experiment_id"
["picontrol","rcp26","rcp45","rcp85"]
$ freva-client databrowser data-count mip_era=mpi-ge variable_id=tas \
frequency=mon --time "2025-01 to 2100-12" --json experiment_id=picontrol \
--host https://www.gems.dkrz.de --flavour cmip6
2
$ freva-client databrowser data-search mip_era=mpi-ge variable_id=tas \
frequency=mon --time "2025-01 to 2100-12" experiment_id=picontrol \
--host https://www.gems.dkrz.de --flavour cmip6
/work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_210001-219912.nc /work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_200001-209912.nc
Search
db_yr.metadata["experiment_id"]
['picontrol', 'rcp26', 'rcp45', 'rcp85']
pcontrol = databrowser(mip_era="mpi-ge",
variable_id="tas",
frequency="mon", time="2025-01 to 2100-12", experiment_id="picontrol",
host="https://www.gems.dkrz.de", flavour="cmip6")
len(pcontrol)
2
for num, file in enumerate(pcontrol):
print(file)
if num > 1:
break
/work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_210001-219912.nc /work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_200001-209912.nc
Search
$ freva-client databrowser metadata-search file=/work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_210001-219912.nc --host=www.gems.dkrz.de
ensemble: r001i1850p3 experiment: picontrol institute: mpi-m model: mpi-esm product: output1 project: mpi-ge realm: atmos time_aggregation: mean time_frequency: mon variable: tas
Search
db.metadata_search(
file="/work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_210001-219912.nc",
host="www.gems.dkrz.de"
)
{'ensemble': ['r001i1850p3'],
'experiment': ['picontrol'],
'institute': ['mpi-m'],
'model': ['mpi-esm'],
'product': ['output1'],
'project': ['mpi-ge'],
'realm': ['atmos'],
'time_aggregation': ['mean'],
'time_frequency': ['mon'],
'variable': ['tas']}
Search
freva-client databrowser metadata-search mip_era=mpi-ge variable_id=tas \
frequency=mon --time="2025-01 to 2100-12" experiment_id='!picontrol' --json \
--host https://www.gems.dkrz.de --flavour cmip6| jq -rc '.experiment_id| join(" ")'
rcp26 rcp45 rcp85
Search
db = databrowser(mip_era="mpi-ge", variable_id="tas",
frequency="mon", time="2025-01 to 2100-12",
experiment_id="!picontrol", flavour="cmip6",
host="https://www.gems.dkrz.de"
)
db.metadata["experiment_id"]
['rcp26', 'rcp45', 'rcp85']
Search
temp_dir=$(mktemp -d --suffix cdo)
for exp in $experiments ;do
outlist=()
# Let's get only the first 5 ensemble member for brevity
members=$(freva-client databrowser metadata-search \
project=mpi-ge variable=tas time_frequency=mon --time="2025-01 to 2100-12" experiment="$exp" --json |
jq -r '.ensemble | unique | .[:5] | join(" ")')
for ens in $members;do
echo -ne "Reading data and calculating TS for experiment $exp in ens: $ens\r"
files=$(freva-client databrowser data-search project=mpi-ge variable=tas time_frequency=mon --time="2025-01 to 2100-12" experiment=$exp ensemble=$ens realm=atmos)
outfile="$temp_dir/tas_mean_${exp}_${ens}.nc"
cdo -s fldmean -mergetime $files "$outfile"
outlist+=("$outfile")
done
cdo mergetime "${outlist[@]}" "$temp_dir/tas_ensemble_${exp}.nc"
done
cdo mergetime $temp_dir/tas_ensemble_*.nc tas_all_experiments.nc
cdo mergetime: Processed 5640 values from 5 variables over 5640 timesteps [0.03s 25MB] cdo mergetime: Processed 5640 values from 5 variables over 5640 timesteps [0.03s 25MB] cdo mergetime: Processed 5640 values from 5 variables over 5640 timesteps [0.03s 25MB]
Search
import matplotlib.pyplot as plt
colors = plt.rcParams["axes.prop_cycle"].by_key()["color"]
plt.figure(figsize=(12, 5))
for i, exp in enumerate(data.experiment.values):
ts = data.sel(experiment=exp).resample(time="1YE").mean()
ts_min = ts.min(dim="ensemble").squeeze()
ts_max = ts.max(dim="ensemble").squeeze()
ts_mean = ts.mean(dim="ensemble").squeeze()
plt.fill_between(
ts.time,
ts_min,
ts_max,
color=colors[i % len(colors)],
alpha=0.2,
label=None,
)
plt.plot(
ts.time,
ts_mean,
color=colors[i % len(colors)],
linewidth=2,
label=f"{exp}"
)
plt.title("Ensemble Mean and Spread for Each Experiment")
plt.xlabel("Time")
plt.ylabel(f"{data.attrs['long_name']} [{data.attrs['units']}]")
plt.legend()
plt.grid(True)
plt.tight_layout()
plt.show()
.
Search
Find data via STAC Browser¶
Cataloging Data (Reusablity)
$ freva-client databrowser intake-catalogue --host www.gems.dkrz.de --flavour cmip6 \
mip_era=mpi-ge frequency=mon variable_id=tas --time "2025-01 to 2100-12" \
experiment_id=picontrol
{
"esmcat_version": "0.1.0",
"attributes": [
{
"column_name": "mip_era",
"vocabulary": ""
},
{
"column_name": "activity_id",
"vocabulary": ""
},
{
"column_name": "institution_id",
"vocabulary": ""
},
{
"column_name": "source_id",
"vocabulary": ""
},
{
"column_name": "experiment_id",
"vocabulary": ""
},
{
"column_name": "frequency",
"vocabulary": ""
},
{
"column_name": "realm",
"vocabulary": ""
},
{
"column_name": "variable_id",
"vocabulary": ""
},
{
"column_name": "member_id",
"vocabulary": ""
},
{
"column_name": "table_id",
"vocabulary": ""
},
{
"column_name": "fs_type",
"vocabulary": ""
},
{
"column_name": "grid_label",
"vocabulary": ""
},
{
"column_name": "format",
"vocabulary": ""
}
],
"assets": {
"column_name": "file",
"format_column_name": "format"
},
"id": "freva",
"description": "Catalogue from freva-databrowser v2507.0.0",
"title": "freva-databrowser catalogue",
"last_updated": "2025-08-05T14:06:40.039213",
"aggregation_control": {
"variable_column_name": "variable_id",
"groupby_attrs": [],
"aggregations": [
{
"type": "union",
"attribute_name": "mip_era",
"options": {}
},
{
"type": "union",
"attribute_name": "activity_id",
"options": {}
},
{
"type": "union",
"attribute_name": "institution_id",
"options": {}
},
{
"type": "union",
"attribute_name": "source_id",
"options": {}
},
{
"type": "union",
"attribute_name": "experiment_id",
"options": {}
},
{
"type": "union",
"attribute_name": "frequency",
"options": {}
},
{
"type": "union",
"attribute_name": "realm",
"options": {}
},
{
"type": "union",
"attribute_name": "variable_id",
"options": {}
},
{
"type": "union",
"attribute_name": "member_id",
"options": {}
},
{
"type": "union",
"attribute_name": "table_id",
"options": {}
},
{
"type": "union",
"attribute_name": "fs_type",
"options": {}
},
{
"type": "union",
"attribute_name": "grid_label",
"options": {}
},
{
"type": "union",
"attribute_name": "format",
"options": {}
}
]
}
,
"catalog_dict": [
{
"file": "/work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_210001-219912.nc",
"project": "MPI-GE",
"product": "output1",
"institute": "MPI-M",
"model": "MPI-ESM",
"experiment": "piControl",
"time_frequency": "mon",
"realm": "atmos",
"variable": "tas",
"ensemble": "r001i1850p3",
"cmor_table": "Amon",
"fs_type": "posix",
"grid_label": "gn",
"format": "nc"
},
{
"file": "/work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/tas/r001i1850p3/v20190123/tas_Amon_MPI-ESM_piControl_r001i1850p3_200001-209912.nc",
"project": "MPI-GE",
"product": "output1",
"institute": "MPI-M",
"model": "MPI-ESM",
"experiment": "piControl",
"time_frequency": "mon",
"realm": "atmos",
"variable": "tas",
"ensemble": "r001i1850p3",
"cmor_table": "Amon",
"fs_type": "posix",
"grid_label": "gn",
"format": "nc"
},
{
"file": "/work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/pr/r001i1850p3/v20190123/pr_Amon_MPI-ESM_piControl_r001i1850p3_210001-219912.nc",
"project": "MPI-GE",
"product": "output1",
"institute": "MPI-M",
"model": "MPI-ESM",
"experiment": "piControl",
"time_frequency": "mon",
"realm": "atmos",
"variable": "pr",
"ensemble": "r001i1850p3",
"cmor_table": "Amon",
"fs_type": "posix",
"grid_label": "gn",
"format": "nc"
},
{
"file": "/work/mh1007/CMOR/MPI-GE/output1/MPI-M/MPI-ESM/piControl/mon/atmos/pr/r001i1850p3/v20190123/pr_Amon_MPI-ESM_piControl_r001i1850p3_200001-209912.nc",
"project": "MPI-GE",
"product": "output1",
"institute": "MPI-M",
"model": "MPI-ESM",
"experiment": "piControl",
"time_frequency": "mon",
"realm": "atmos",
"variable": "pr",
"ensemble": "r001i1850p3",
"cmor_table": "Amon",
"fs_type": "posix",
"grid_label": "gn",
"format": "nc"
}
]
}
.
Catalog
db = databrowser(mip_era="mpi-ge", variable_id="tas",
frequency="mon", time="2025-01 to 2100-12",
experiment_id="picontrol", flavour="cmip6",
host="https://www.gems.dkrz.de"
)
db.intake_catalogue()
freva catalog with 2 dataset(s) from 2 asset(s):
| unique | |
|---|---|
| file | 2 |
| project | 1 |
| product | 1 |
| institute | 1 |
| model | 1 |
| experiment | 1 |
| time_frequency | 1 |
| realm | 1 |
| variable | 1 |
| ensemble | 1 |
| cmor_table | 1 |
| fs_type | 1 |
| grid_label | 1 |
| format | 1 |
| derived_variable_id | 0 |
Catalog
$ freva-client databrowser stac-catalogue --host www.gems.dkrz.de --flavour cmip6 \
mip_era=mpi-ge frequency=mon variable_id=tas --time "2025-01 to 2100-12" \
experiment_id=picontrol
Downloading the STAC catalog started ... STAC catalog saved to: /Users/mo/dev/20250805/Talks/talks/DataSearchWorkshop2025/stac-catalog-Dataset-cmip6-ca87bec5-e66-file.zip (size: 0.02 MB). Or simply download from: https://www.gems.dkrz.de/api/freva-nextgen/databrowser/stac-catalogue/cmip6/file?multi-version=False&mip_era=mpi-ge&frequency=mon&variable_id=pr&variable_id=tas&experiment_id=picontrol&time=2025-01+to+2100-12&time_select=flexible
Catalog
db = databrowser(mip_era="mpi-ge", variable_id="tas",
frequency="mon", time="2025-01 to 2100-12",
experiment_id="picontrol", flavour="cmip6",
host="https://www.gems.dkrz.de"
)
db.stac_catalogue()
Downloading the STAC catalog started ...
'STAC catalog saved to: /Users/mo/dev/20250805/Talks/talks/DataSearchWorkshop2025/stac-catalog-Dataset-cmip6-ec013869-2c5-file.zip (size: 0.01 MB). Or simply download from: https://www.gems.dkrz.de/api/freva-nextgen/databrowser/stac-catalogue/cmip6/file?multi-version=False&mip_era=mpi-ge&variable_id=tas&frequency=mon&experiment_id=picontrol&time=2025-01+to+2100-12&time_select=flexible'
Catalog
Customize Dataset (Interoperablity)
To continue this section please open the Tutorial-py-userdata.ipynb or Tutorial-shell-userdata.ipynb, based on your desire, and then from kernel environment list, please choose, DA Workshop (python) or DA Workshop (shell) again based on your preferences.
Custom
USER=$(whoami)
OUTFILE="dummy_sst_${USER}.nc"
GRIDFILE="$(mktemp)"
cat > "$GRIDFILE" << EOF
gridtype = lonlat
xsize = 171
ysize = 121
xfirst = 120
xinc = 1
yfirst = -30
yinc = 0.5
xname = lon
yname = lat
xunits = degrees_east
yunits = degrees_north
EOF
cdo -f nc const,0,"$GRIDFILE" base.nc
cdo -f nc \
-setcalendar,standard \
-settaxis,2025-01-01,00:00:00,1month \
-expr,'sst=28-0.006*(clon(const)-230)' \
-setattribute,sst@long_name="Idealized Pacific SST" \
-setattribute,sst@units="°C" \
base.nc "$OUTFILE"
rm -f base.nc "$GRIDFILE"
echo "Wrote $OUTFILE → exists? $( [[ -f $OUTFILE ]] && echo yes || echo no )"
Wrote dummy_sst_k202187.nc → exists? yes
Custom
import numpy as np, xarray as xr, os
from getpass import getuser
from freva_client import databrowser, authenticate
from pathlib import Path
time = np.arange("2025-01","2026-01",dtype="datetime64[M]")
lat, lon = np.linspace(-30,30,121), np.linspace(120,290,171)
da = (xr.DataArray(28 - 0.006*(lon-230), dims=("lon",), coords={"lon":lon})
.expand_dims(time=time, lat=lat))
da.name = "sst"; da.attrs.update(long_name="Idealized Pacific SST", units="°C")
da.to_netcdf(f"dummy_sst_{getuser()}.nc")
print(f"check if dummy_sst_{getuser()}.nc exists: {os.path.exists(f'dummy_sst_{getuser()}.nc')}")
check if dummy_sst_mo.nc exists: True
Custom
import matplotlib.pyplot as plt, cartopy.crs as ccrs, cartopy.feature as cfeature
fig, ax = plt.subplots(figsize=(8,4), subplot_kw=dict(projection=ccrs.PlateCarree(180)))
da.mean("time").plot.pcolormesh(ax=ax, transform=ccrs.PlateCarree(), cmap="coolwarm", add_colorbar=True)
ax.add_feature(cfeature.LAND, facecolor="white", zorder=2); ax.coastlines(zorder=3); ax.add_feature(cfeature.BORDERS, linestyle=":", zorder=3)
plt.title("Pacific SST"); plt.tight_layout(); plt.show()
Auth
cat > ~/.freva-access-token.json << 'EOF'
EOF
token = """ """
_ = (Path.home() / ".freva-access-token.json").write_text(token)
Custom
global_attributes = {"project": "userdata", "product": "stats", "model": "IFS", "experiment": "ETCCDI", "realm": "atmos"}
databrowser.userdata(
action="add",
userdata_items=[f"{os.getcwd()}/dummy_sst_{getuser()}.nc"],
metadata=global_attributes,
)
1 have been successfully added to the databrowser. 0 files were duplicates and not added.
global_attributes = {"project": "userdata", "product": "stats", "model": "IFS", "experiment": "ETCCDI", "realm": "atmos"}
databrowser.userdata(
action="delete",
metadata=global_attributes,
)
User data deleted successfully
Custom
$ freva-client databrowser user-data add --path dummy_sst_mo.nc --facet project=userdata --facet product=stats --facet model=IFS --facet experiment=ETCCDI --facet realm=atmos --token-file ~/.freva-access-token.json
1 have been successfully added to the databrowser. 0 files were duplicates and not added.
$ freva-client databrowser user-data delete --search-key project=userdata --search-key product=stats --search-key model=IFS --search-key experiment=ETCCDI --search-key realm=atmos --token-file ~/.freva-access-token.json
User data deleted successfully
Custom
S3 Access (Accessibility)
S3
File vs Object Storage
File Storage¶
- Hierarchical structure (tree)
- Directory with files and other directories
- Easy to rename and move items around
- Not identified by name
- Change references
- System-centric, not designed to be share
S3
File vs Object Storage
Object Storage¶
- Flat structure (key-value)
- Items are identified by a key/name
- Content is accessed providing key/name
- Not easy to move items around!
- Moving is renaming
- Modify key
S3
Local vs Remote
Local storage should be:
- Fast
- Reliable
- Performant!
Remote storage should be:
- Simple & common language
- Permissive
- Highly available!
S3
HTTP aka internet language!?¶
Stands for Hypertext Transfer Protocol. There are many protocols...
- simple
- very generic
- very flexible
- famous!
Client makes a request, server replies
NOTE: IP the actual and literal Internel Protocol
S3
Replies¶
- Status Code -(
404 Not Found) - Headers - metadata, options (
Content-Type,...) - Body - data (empty, raw bytes, text,... )
S3
Object Store APIs¶
Providers expose application programming interface (API) which might not be (fully) interchangable. E.g. list objects:
GET /?delimiter={delimiter}&encoding-type={enctype}&marker=....&prefix={prefix}
GET /v1/{account}/{container}
GET /v1/b/{bucket}/o/
GET /{container}?restype=container&comp=list
S3
S3 - Simple Storage Service¶
Created by AWS
- S3 API is public (partially)
S3
S3 Concepts¶
- Bucket - The container of the objects
- Objects - The actual data
- Regions (advanced) - Location of the data
- Authentication
Directories can be emulated by empty objects terminated with
/
S3
S3 - Authentication¶
- A bucket may not be publicly available.
- Key-Secret pairs sign the request
- May expire automatically
- May be valid indefinetly
- Create temporarly link to share or receive data
S3
Exploring data on S3¶
- Command line interfaces (CLIs)
- Software Development Kits (SDKs)
- Libraries
#conda install awscli ||pip install awscli
aws s3 ls s3://ncar-cesm2-lens/ --no-sign-request
alias awss3='aws s3 --no-sign-request'
awss3 cp --recursive s3://ncar-cesm2-lens/atm/static/grid.zarr /tmp/grid.zarr
s3:// is a comodity:
s3://ncar-cesm2-lensresolves tohttps://s3.us-west-2.amazonaws.com/ncar-cesm2-lens- Behaviour dependens on the
AWS_*environment variables
Appending
/to the S3 resource may produce different results
module load minio-client
mc alias set aws https://s3.amazonaws.com "" "" # empty keys = no authentication
mc ls aws/ncar-cesm2-lens/ice/static/grid.zarr
mc alias set dkrz https://s3.eu-dkrz-1.dkrz.cloud "" "" # no authentication -> read-only!!!
mc ls dkrz/freva
#mc cp /scratch/${USER:0:1}/${USER}/path/to/my/file.txt dkrz/freva/workshop/${USER}/myfile.txt
S3
The usual suspects¶
dataset_url = 's3://ncar-cesm2-lens/ocn/static/grid.zarr'
s3_opts = {'anon': True }
# s3_minio_opt = { # From playground: https://docs.min.io/community/minio-object-store/developers/python/minio-py.html#id3
# 'key': 'Q3AM3UQ867SPQQA43P2F',
# 'secret': 'zuf+tfteSlswRu7BJ86wekitnifILbZam1KYY3TG',
# 'client_kwargs':{'endpoint_url': 'https://play.min.io'}
# }
import xarray as xr
ds = xr.open_dataset(dataset_url, engine='zarr', storage_options=s3_opts)
import zarr
zarr.open(dataset_url, storage_options=s3_opts)
S3
NetCDF on S3 (classic, nc4/hdf5)¶
import s3fs
s3 = s3fs.S3FileSystem(endpoint_url="https://s3.eu-dkrz-1.dkrz.cloud", anon=True)
with s3.open('s3://freva/workshop/tas.nc') as f:
ds = xr.open_dataset(f)
We cannot write directly via xarray. The reason is that Netcdf jumps back and forth while writing the file, which is not supported by S3.
Instead, we can write to a file or memory buffer which we then upload to S3.
In Levante custom builds of libnetcdf and cdo can work with data on S3
Very unstable!!!
/fastdata/freva/opt/bin/ncdump -h https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/tas.nc#mode=s3,bytes
/fastdata/freva/opt/bin/cdo sinfo https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/tas.nc#mode=s3,bytes
S3 Handson
Handson - S3 Data Access¶
For this part we have a single python the notebook.
Assuming you previously executed the setup stetps:
$ module load clint gems
$ da-workshop-setup
Open the Tutorial-py-s3.ipynb and then from kernel environment list, please choose, DA Workshop (python).
Let's then start! :)
S3 Handson
strip_dict(databrowser.metadata_search(fs_type="s3"))
{'ensemble': ['r1i1p1f1'],
'experiment': ['historical', 'ssp370'],
'institute': ['ncar'],
'model': ['cesm2-le'],
'product': ['output'],
'project': ['cesm2-le'],
'realm': ['atm', 'ice', 'lnd', '...'],
'time_aggregation': ['mean'],
'time_frequency': ['daily', 'monthly', 'static'],
'variable': ['aice', 'aice_d', 'dic', '...']}
S3 Handson
search_keys = {
'fs_type': 's3',
'project':'cesm2-le',
'experiment': 'historical',
'realm': 'atm',
'time_frequency':'monthly'
}
'ts' in databrowser.metadata_search(**search_keys)['variable']
True
db = databrowser(variable='ts', **search_keys)
list(db)
['s3://ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-TS.zarr', 's3://ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-TS.zarr']
S3 Handson
Let's quickly check how to open the first dataset with xarray¶
Since all are zarr datasets we can use xr.open_zarr or xr.open_dataset(engine='zarr').
Because the data is public, we do not need credentials to open the data. We specify anon (anonymous) in order for xarray to not sign the requests.
If data is not public we will get 403 Forbidden
S3 Handson
import xarray as xr
xr.open_zarr(list(db)[0], storage_options={ 'anon':True })
print(xr.open_dataset(list(db)[0], engine='zarr', storage_options={ 'anon':True }))
<xarray.Dataset> Size: 22GB
Dimensions: (member_id: 50, time: 1980, lat: 192, lon: 288, nbnd: 2)
Coordinates:
* lat (lat) float64 2kB -90.0 -89.06 -88.12 ... 88.12 89.06 90.0
* lon (lon) float64 2kB 0.0 1.25 2.5 ... 355.0 356.2 357.5 358.8
* member_id (member_id) <U12 2kB 'r10i1191p1f2' ... 'r9i1171p1f2'
* time (time) object 16kB 1850-01-16 12:00 ... 2014-12-16 12:00:00
time_bnds (time, nbnd) object 32kB ...
Dimensions without coordinates: nbnd
Data variables:
TS (member_id, time, lat, lon) float32 22GB ...
Attributes:
Conventions: CF-1.0
logname: sunseon
model_doi_url: https://doi.org/10.5065/D67H1H0V
source: CAM
time_period_freq: month_1
topography_file: /mnt/lustre/share/CESM/cesm_input/atm/cam/topo/fv...
S3 Handson
from pathlib import Path
s3_opts = { 'anon':True }
time_series = {}
for fileurl in db:
print(f"Opening {fileurl}...")
engine = {'engine':'zarr'} if fileurl.endswith('zarr') else {}
ds = xr.open_dataset(fileurl, **engine, storage_options=s3_opts)
ds = ds.rename({'member_id':'ensemble'})
# Going through all ensembles might take some time
# let's make a cut at 5 member for demo purposes
# Memory usage might spike to ~70GB
members=ds.ensemble[:5]
ds = ds.sel(ensemble=members)
# cesm2LE-historical-cmip6-TS cesm2LE-historical-smbb-TS
dataset_name=Path(fileurl).stem
mean_ts = field_mean(ds["TS"])
mean_ts.attrs['source_dataset'] = fileurl
time_series[dataset_name] = mean_ts
Opening s3://ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-TS.zarr Opening s3://ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-TS.zarr
S3 Handson
Plot the data (same as before)
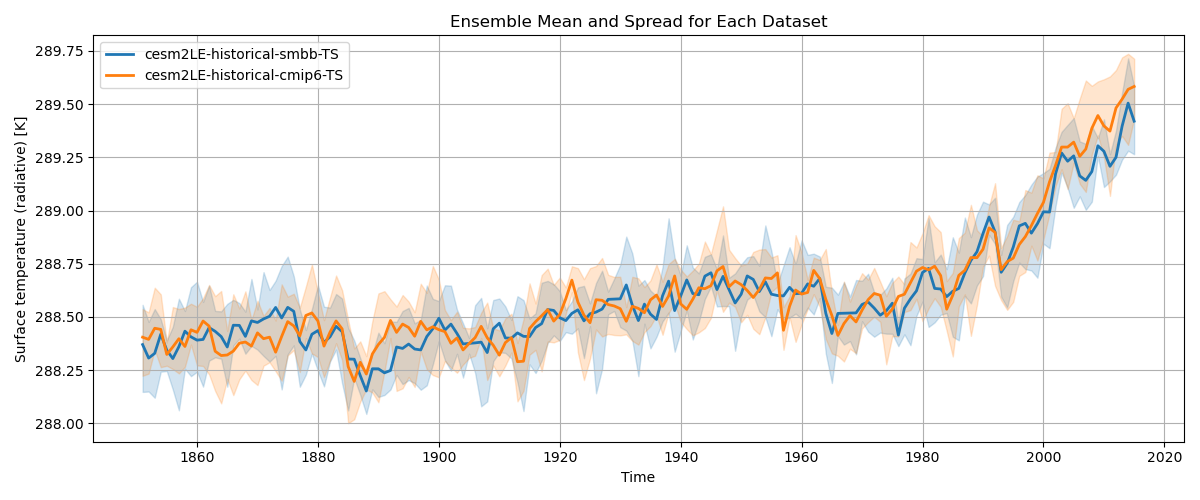
S3 Handson
import matplotlib.pyplot as plt
# Create a color cycle for different experiments
colors = plt.rcParams["axes.prop_cycle"].by_key()["color"]
plot = plt.figure(figsize=(12, 5))
for i, (exp, data) in enumerate(time_series.items()):
ts = data.resample(time="1YE").mean()
ts_min = ts.min(dim="ensemble").squeeze()
ts_max = ts.max(dim="ensemble").squeeze()
ts_mean = ts.mean(dim="ensemble").squeeze()
time_values = np.array(ts.time.values, dtype='datetime64[ns]')
# Plot min–max shading
plt.fill_between(
time_values,
ts_min.values,
ts_max.values,
color=colors[i % len(colors)],
alpha=0.2,
label=None,
)
# Plot mean line
plt.plot(
time_values,
ts_mean.values,
color=colors[i % len(colors)],
linewidth=2,
label=f"{exp}"
)
# Add plot decorations
plt.title("Ensemble Mean and Spread for Each Dataset")
plt.xlabel("Time")
plt.ylabel(f"{data.attrs['long_name']} [{data.attrs['units']}]") # Replace with actual units
plt.legend()
plt.grid(True)
plt.tight_layout()
plt.show()
S3 Handson
S3 Handson
We start by setting up a dictionary with the configuration we will need!
from getpass import getuser # to get USER env variable
USERNAME = getuser()
s3_config = {
'bucket' : 'freva',
'endpoint' :'https://s3.eu-dkrz-1.dkrz.cloud', # DKRZ Minio S3
'prefix' : f'workshop/{USERNAME}', # Avoid users writting object with same prefix
'access_key_id' : "s3handson", # Only valid during the workshop
'secret_access_key' : "s3handson", # Only valid during the workshop
'region' : 'eu-dkrz-1',
}
S3 Handson
Now we create a wrapper on S3 that mimics a local filesystem. This will be important for netcdf
import s3fs
s3 = s3fs.S3FileSystem(
key = s3_config['access_key_id'],
secret = s3_config['secret_access_key'],
client_kwargs = {'endpoint_url': s3_config['endpoint']},
)
full_prefix = s3_config['bucket']+'/'+s3_config['prefix']
testobj = f'{full_prefix}/hi.txt'
s3.write_bytes(testobj, b'Hi!\n')
print(f"Writing data to S3 works! Test it with:\ncurl {s3_config['endpoint']}/{testobj}")
Writing data to S3 works! Test it with: curl https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/k202186/hi.txt
!curl https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/${USER}/hi.txt
Hi!
S3 Handson
Save the figure on S3¶
- Open the object ("file") on S3 where the figure should be saved
- Save the figure into that opened object!
figure_path = f'{full_prefix}/figure-ts-mean.png'
with s3.open(figure_path, 'wb') as f:
plot.savefig(f)
print(f"Open this link to get the image: {s3_config['endpoint']}/{figure_path}")
Open this link to get the image: https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/k202186/figure-ts-mean.png
S3 Handson
Let's now write the data to S3 as both NetCDF and ZARR¶
Since we are writting to an object store we cannot pass an opened file to xr.to_netcdf because it will use seek operations, which are not allowed in S3!
In that case that case simply copy the file. For zarr it's ok because seek is not used.
def write_netcdf(s3_path, dataset:xr.Dataset):
tmp_name = f"/scratch/{USERNAME[0]}/{USERNAME}/{name}.nc"
dataset.to_netcdf(tmp_name, engine='h5netcdf')
## Copy to s3
with s3.open(s3_path,'wb') as s3file:
with open(tmp_name, 'rb') as tmpf:
s3file.write(tmpf.read())
## Check if we can open with Xarray!
with s3.open(s3_path,'rb') as s3file:
xr.open_dataset(s3file, engine='h5netcdf')
## We can now remove the local copy
import os
os.remove(tmp_name)
S3 Handson
With Zarr is enough to pass the S3 configuration in order to have xarray writing the dataset
def write_zarr(store, dataset: xr.Dataset):
import zarr
zarr.config.set(default_zarr_format=2)
import numcodecs
codec = numcodecs.Blosc(shuffle=1, clevel=6)
data_encoding = {}
for var in dataset.variables:
data_encoding[var] = { "compressors": codec}
dataset.to_zarr(store,
mode='w', # OVERWRITES existing data!!
encoding=data_encoding,
consolidated=True, # consolidate metadata for fast access
storage_options= { # We cannot use the anonymous mode anymore
'key':s3_config['access_key_id'],
'secret':s3_config['secret_access_key'],
'client_kwargs':{
'endpoint_url': s3_config['endpoint']
},
})
S3 Handson
def write_dataset_to_s3(name:str, dataset:xr.Dataset, file_format='nc'):
_supported_types = ('nc','zarr')
file_format = file_format.replace('.','')
if file_format not in _supported_types:
raise Exception('Unsuported file format, use one of')
s3_path = f'{full_prefix}/{name}.{file_format}'
if file_format == 'nc':
write_netcdf(s3_path, dataset)
elif file_format == 'zarr':
write_zarr(f's3://{s3_path}', dataset)
netcdf_mode= 'zarr' if file_format == 'zarr' else 'bytes'
print(f"Try running:\n\t" \
f"/fastdata/freva/opt/bin/ncdump -h {s3_config['endpoint']}/{s3_path}#mode=s3,{netcdf_mode}\n\t"\
f"/fastdata/freva/opt/bin/cdo sinfo {s3_config['endpoint']}/{s3_path}#mode=s3,{netcdf_mode}")
S3 Handson
We save the mean temperature array in S3 as both Zarr and NetCDF
for name, data in time_series.items():
## for CDO it is important that time is the first dimension
## also it cannot have indexers of type string
ensemble = data.ensemble.values
ensemble_id = np.arange(len(ensemble))
dataset = data.to_dataset() \
.transpose('time',...) \
.assign_coords(ensemble_id=('ensemble',ensemble_id)) \
.swap_dims({'ensemble':'ensemble_id'}) \
.reset_coords(drop=True)\
.assign_attrs(ensembles= ', '.join(data.ensemble.values))
write_dataset_to_s3(name, dataset, file_format='nc')
write_dataset_to_s3(name, dataset, file_format='zarr')
public_url = f"https://eu-dkrz-1.dkrz.cloud/browser/{s3_config['bucket']}/{s3_config['prefix']}"
f"{s3_config['endpoint']}/browser/{s3_config['bucket']}/{s3_config['prefix']}"
print(f"Browser the data in {public_url}")
Try running: /fastdata/freva/opt/bin/ncdump -h https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/k202186/cesm2LE-historical-smbb-TS.nc#mode=s3,bytes ... Browser the data in https://eu-dkrz-1.dkrz.cloud/browser/freva/workshop/k202186
S3 Handson
As the output suggests, we should be able to see the contents of the bucket:
Also, note the diference between endpoints:
eu-dkrz-1.dkrz.cloudis for the web user interfaces3.eu-dkrz-1.dkrz.cloudis for the data access
With a custom netcdf build (/fastdata/freva/opt), we can pass an URL with fragment mode:
s3for any link that is an S3 URLbytesfor raw netcdf datazarrfor zarr data
E.g.:
export PATH=/fastdata/freva/opt/bin/:${PATH}
ncdump -h https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/tas.nc#mode=s3,bytes
ncdump -h https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/tas.zarr#mode=s3,zarr
S3 Handson
!/fastdata/freva/opt/bin/ncdump -h https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/$USER/cesm2LE-historical-smbb-TS.nc#mode=s3,bytes
!/fastdata/freva/opt/bin/cdo sinfo https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/$USER/cesm2LE-historical-smbb-TS.nc#mode=s3,bytes
netcdf cesm2LE-historical-smbb-TS {
dimensions:
time = 1980 ;
ensemble_id = 5 ;
variables:
double time(time) ;
time:_FillValue = NaN ;
string time:units = "days since 1850-01-01" ;
string time:calendar = "noleap" ;
double TS(time, ensemble_id) ;
TS:_FillValue = NaN ;
string TS:cell_methods = "time: mean" ;
string TS:long_name = "Surface temperature (radiative)" ;
string TS:units = "K" ;
string TS:source_dataset = "s3://ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-TS.zarr" ;
int64 ensemble_id(ensemble_id) ;
// global attributes:
string :ensembles = "r10i1191p1f2, r11i1231p1f2, r11i1251p1f2, r11i1281p1f2, r11i1301p1f2" ;
}
File format : NetCDF4
-1 : Institut Source T Steptype Levels Num Points Num Dtype : Parameter ID
1 : unknown unknown v instant 1 1 5 1 F64 : -1
Grid coordinates :
1 : generic : points=5
ensemble_id : 0 to 4 by 1
Vertical coordinates :
1 : surface : levels=1
Time coordinate :
time : 1980 steps
RefTime = 1850-01-01 00:00:00 Units = days Calendar = 365_day
YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss
1850-01-16 12:00:00 1850-02-15 00:00:00 1850-03-16 12:00:00 1850-04-16 00:00:00
1850-05-16 12:00:00 1850-06-16 00:00:00 1850-07-16 12:00:00 1850-08-16 12:00:00
1850-09-16 00:00:00 1850-10-16 12:00:00 1850-11-16 00:00:00 1850-12-16 12:00:00
1851-01-16 12:00:00 1851-02-15 00:00:00 1851-03-16 12:00:00 1851-04-16 00:00:00
1851-05-16 12:00:00 1851-06-16 00:00:00 1851-07-16 12:00:00 1851-08-16 12:00:00
1851-09-16 00:00:00 1851-10-16 12:00:00 1851-11-16 00:00:00 1851-12-16 12:00:00
1852-01-16 12:00:00 1852-02-15 00:00:00 1852-03-16 12:00:00 1852-04-16 00:00:00
1852-05-16 12:00:00 1852-06-16 00:00:00 1852-07-16 12:00:00 1852-08-16 12:00:00
1852-09-16 00:00:00 1852-10-16 12:00:00 1852-11-16 00:00:00 1852-12-16 12:00:00
1853-01-16 12:00:00 1853-02-15 00:00:00 1853-03-16 12:00:00 1853-04-16 00:00:00
1853-05-16 12:00:00 1853-06-16 00:00:00 1853-07-16 12:00:00 1853-08-16 12:00:00
1853-09-16 00:00:00 1853-10-16 12:00:00 1853-11-16 00:00:00 1853-12-16 12:00:00
1854-01-16 12:00:00 1854-02-15 00:00:00 1854-03-16 12:00:00 1854-04-16 00:00:00
1854-05-16 12:00:00 1854-06-16 00:00:00 1854-07-16 12:00:00 1854-08-16 12:00:00
1854-09-16 00:00:00 1854-10-16 12:00:00 1854-11-16 00:00:00 1854-12-16 12:00:00
................................................................................
................................................................................
..........
2010-01-16 12:00:00 2010-02-15 00:00:00 2010-03-16 12:00:00 2010-04-16 00:00:00
2010-05-16 12:00:00 2010-06-16 00:00:00 2010-07-16 12:00:00 2010-08-16 12:00:00
2010-09-16 00:00:00 2010-10-16 12:00:00 2010-11-16 00:00:00 2010-12-16 12:00:00
2011-01-16 12:00:00 2011-02-15 00:00:00 2011-03-16 12:00:00 2011-04-16 00:00:00
2011-05-16 12:00:00 2011-06-16 00:00:00 2011-07-16 12:00:00 2011-08-16 12:00:00
2011-09-16 00:00:00 2011-10-16 12:00:00 2011-11-16 00:00:00 2011-12-16 12:00:00
2012-01-16 12:00:00 2012-02-15 00:00:00 2012-03-16 12:00:00 2012-04-16 00:00:00
2012-05-16 12:00:00 2012-06-16 00:00:00 2012-07-16 12:00:00 2012-08-16 12:00:00
2012-09-16 00:00:00 2012-10-16 12:00:00 2012-11-16 00:00:00 2012-12-16 12:00:00
2013-01-16 12:00:00 2013-02-15 00:00:00 2013-03-16 12:00:00 2013-04-16 00:00:00
2013-05-16 12:00:00 2013-06-16 00:00:00 2013-07-16 12:00:00 2013-08-16 12:00:00
2013-09-16 00:00:00 2013-10-16 12:00:00 2013-11-16 00:00:00 2013-12-16 12:00:00
2014-01-16 12:00:00 2014-02-15 00:00:00 2014-03-16 12:00:00 2014-04-16 00:00:00
2014-05-16 12:00:00 2014-06-16 00:00:00 2014-07-16 12:00:00 2014-08-16 12:00:00
2014-09-16 00:00:00 2014-10-16 12:00:00 2014-11-16 00:00:00 2014-12-16 12:00:00
cdo sinfo: Processed 1 variable over 1980 timesteps [0.13s 3000MB]
S3 Handson
!/fastdata/freva/opt/bin/ncdump -h https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/$USER/cesm2LE-historical-smbb-TS.zarr#mode=s3,zarr
!/fastdata/freva/opt/bin/cdo sinfo https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/$USER/cesm2LE-historical-smbb-TS.zarr#mode=s3,zarr
netcdf cesm2LE-historical-smbb-TS {
dimensions:
time = 1980 ;
ensemble_id = 5 ;
variables:
double TS(time, ensemble_id) ;
TS:_FillValue = NaN ;
TS:cell_methods = "time: mean" ;
TS:long_name = "Surface temperature (radiative)" ;
TS:units = "K" ;
TS:source_dataset = "s3://ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-TS.zarr" ;
int64 time(time) ;
time:units = "hours since 1850-01-16 12:00:00.000000" ;
time:calendar = "noleap" ;
int64 ensemble_id(ensemble_id) ;
// global attributes:
:ensembles = "r10i1191p1f2, r11i1231p1f2, r11i1251p1f2, r11i1281p1f2, r11i1301p1f2" ;
}
File format : NCZarr filter
-1 : Institut Source T Steptype Levels Num Points Num Dtype : Parameter ID
1 : unknown unknown v instant 1 1 5 1 F64f : -1
Grid coordinates :
1 : generic : points=5
ensemble_id : 0 to 4 by 1
Vertical coordinates :
1 : surface : levels=1
Time coordinate :
time : 1980 steps
RefTime = 1850-01-16 12:00:00 Units = hours Calendar = 365_day
YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss
1850-01-16 12:00:00 1850-02-15 00:00:00 1850-03-16 12:00:00 1850-04-16 00:00:00
1850-05-16 12:00:00 1850-06-16 00:00:00 1850-07-16 12:00:00 1850-08-16 12:00:00
1850-09-16 00:00:00 1850-10-16 12:00:00 1850-11-16 00:00:00 1850-12-16 12:00:00
1851-01-16 12:00:00 1851-02-15 00:00:00 1851-03-16 12:00:00 1851-04-16 00:00:00
1851-05-16 12:00:00 1851-06-16 00:00:00 1851-07-16 12:00:00 1851-08-16 12:00:00
1851-09-16 00:00:00 1851-10-16 12:00:00 1851-11-16 00:00:00 1851-12-16 12:00:00
1852-01-16 12:00:00 1852-02-15 00:00:00 1852-03-16 12:00:00 1852-04-16 00:00:00
1852-05-16 12:00:00 1852-06-16 00:00:00 1852-07-16 12:00:00 1852-08-16 12:00:00
1852-09-16 00:00:00 1852-10-16 12:00:00 1852-11-16 00:00:00 1852-12-16 12:00:00
1853-01-16 12:00:00 1853-02-15 00:00:00 1853-03-16 12:00:00 1853-04-16 00:00:00
1853-05-16 12:00:00 1853-06-16 00:00:00 1853-07-16 12:00:00 1853-08-16 12:00:00
1853-09-16 00:00:00 1853-10-16 12:00:00 1853-11-16 00:00:00 1853-12-16 12:00:00
1854-01-16 12:00:00 1854-02-15 00:00:00 1854-03-16 12:00:00 1854-04-16 00:00:00
1854-05-16 12:00:00 1854-06-16 00:00:00 1854-07-16 12:00:00 1854-08-16 12:00:00
1854-09-16 00:00:00 1854-10-16 12:00:00 1854-11-16 00:00:00 1854-12-16 12:00:00
................................................................................
................................................................................
..........
2010-01-16 12:00:00 2010-02-15 00:00:00 2010-03-16 12:00:00 2010-04-16 00:00:00
2010-05-16 12:00:00 2010-06-16 00:00:00 2010-07-16 12:00:00 2010-08-16 12:00:00
2010-09-16 00:00:00 2010-10-16 12:00:00 2010-11-16 00:00:00 2010-12-16 12:00:00
2011-01-16 12:00:00 2011-02-15 00:00:00 2011-03-16 12:00:00 2011-04-16 00:00:00
2011-05-16 12:00:00 2011-06-16 00:00:00 2011-07-16 12:00:00 2011-08-16 12:00:00
2011-09-16 00:00:00 2011-10-16 12:00:00 2011-11-16 00:00:00 2011-12-16 12:00:00
2012-01-16 12:00:00 2012-02-15 00:00:00 2012-03-16 12:00:00 2012-04-16 00:00:00
2012-05-16 12:00:00 2012-06-16 00:00:00 2012-07-16 12:00:00 2012-08-16 12:00:00
2012-09-16 00:00:00 2012-10-16 12:00:00 2012-11-16 00:00:00 2012-12-16 12:00:00
2013-01-16 12:00:00 2013-02-15 00:00:00 2013-03-16 12:00:00 2013-04-16 00:00:00
2013-05-16 12:00:00 2013-06-16 00:00:00 2013-07-16 12:00:00 2013-08-16 12:00:00
2013-09-16 00:00:00 2013-10-16 12:00:00 2013-11-16 00:00:00 2013-12-16 12:00:00
2014-01-16 12:00:00 2014-02-15 00:00:00 2014-03-16 12:00:00 2014-04-16 00:00:00
2014-05-16 12:00:00 2014-06-16 00:00:00 2014-07-16 12:00:00 2014-08-16 12:00:00
2014-09-16 00:00:00 2014-10-16 12:00:00 2014-11-16 00:00:00 2014-12-16 12:00:00
cdo sinfo: Processed 1 variable over 1980 timesteps [0.10s 3000MB]
S3 Handson
print("Opening")
for name in time_series.keys():
file = f'https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/{USERNAME}/{name}'
print(f"{file}.nc\n{file}.zarr")
xr.open_dataset(f'{file}.nc', engine='h5netcdf')
xr.open_zarr(f'{file}.zarr')
print('All output datasets opened!')
Opening https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/k202186/cesm2LE-historical-smbb-TS.nc https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/k202186/cesm2LE-historical-smbb-TS.zarr https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/k202186/cesm2LE-historical-cmip6-TS.nc https://s3.eu-dkrz-1.dkrz.cloud/freva/workshop/k202186/cesm2LE-historical-cmip6-TS.zarr All output datasets opened!
After this walkthrough, we believe FREVA is FAIR enough!
What do you think?
Wrap-up
What we couldn't cover today¶
- Add your Data Analysis code:
- Freva helps you to create UI's for Web, CLI and python
- Reproduciple
- Shareable
- AI tools such as frevaGPT (sneak peek via: gems.dkrz.de/chatbot)
Wrap-up
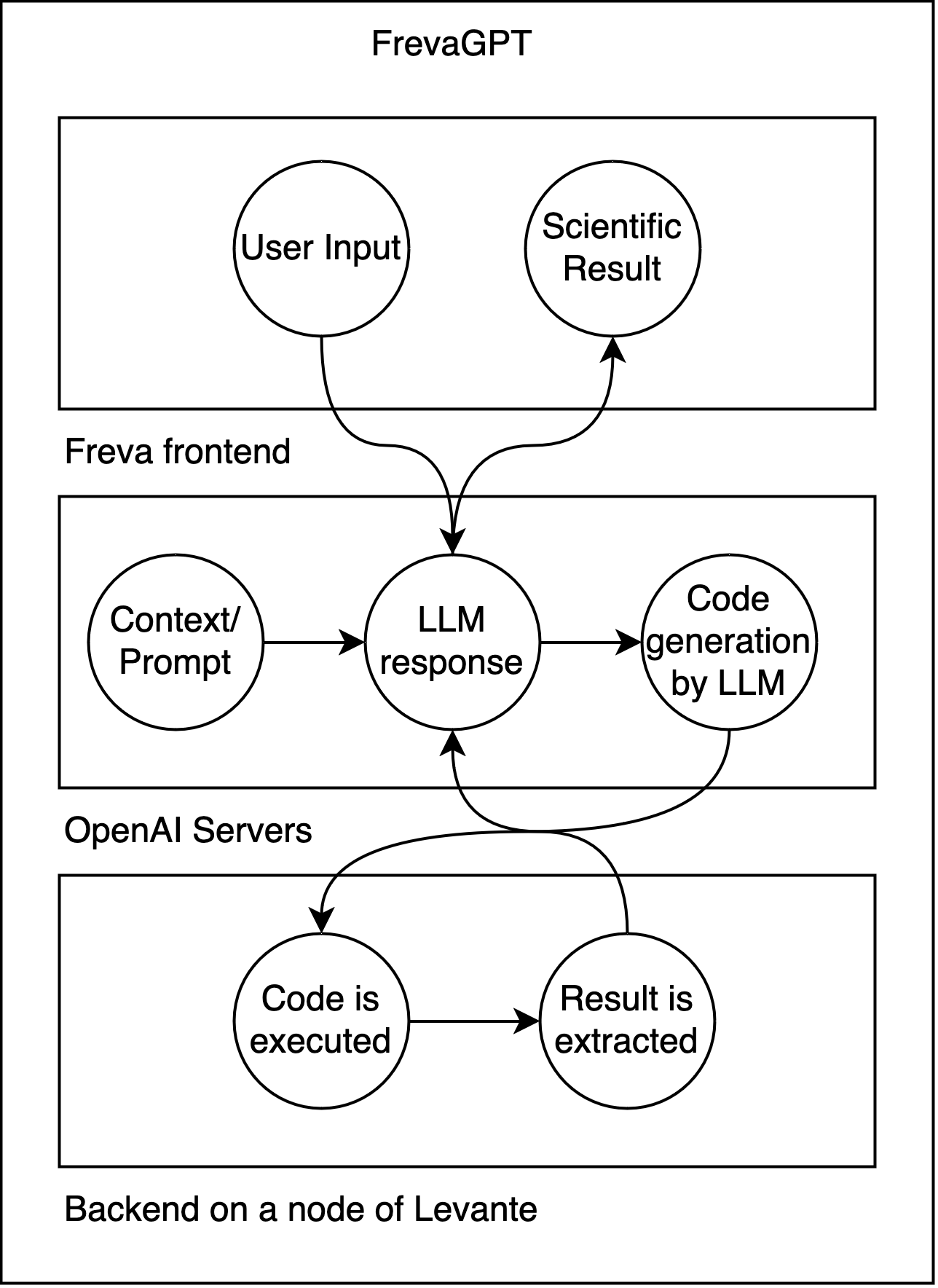
What is FrevaGPT? (pre-Beta!)¶
🤖 FrevaGPT is an AI assistant built into the Freva ecosystem. It uses large language models (LLMs) like GPT-4 alongside a live Python interpreter.
⚙️ It runs code directly on hybrid CPU/GPU nodes at DKRZ's Levante, operating on real data!
⚙️ It is also integrated with JupyterAI frontend for extended functionality with jupyterhub.
🚀 It serves as a powerful stepping stone to explore and analysis data using Freva.
➤ You can currently try it at: https://gems.dkrz.de/chatbot/
Useful Info
Tutorial-{I,II,III}-*.ipynb| hostname | command (levante) | obs |
|---|---|---|
| https://gems.dkrz.de | module load clint gems |
only data browser |
| https://freva.dkrz.de | module load clint freva |
with plugins ⚠️Need to add batch scheduling info in Extra scheduler options⚠️ |